This is a conceptual overview over the modeling functionality in
EpiSewer. See vignette("model-definition") for
a mathematical definition of the underlying generative model, and
vignette("detailed-example") for an example vignette.
Modules
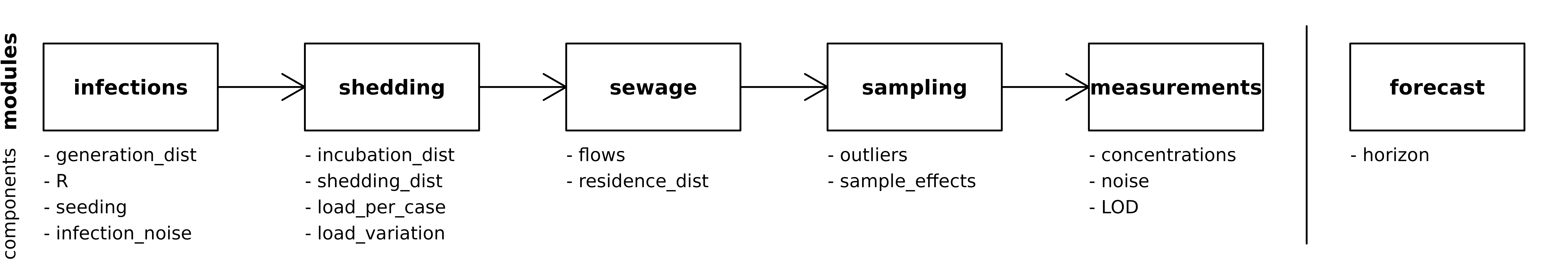
EpiSewer uses 5 different modules to describe the data
generating process behind the wastewater measurements:
infections, shedding, sewage,
sampling, and measurements. There is a 6th
module to specify forecast functionality. Each of these
modules consists of a number of module components, as shown below.
The modules are defined using their corresponding module function,
i.e. by calling model_infections(),
model_shedding(), model_sewage(),
model_sampling(), model_measurements(), or
model_forecast().
Modeling functions
Components in a module can be specified using suitable modeling functions. There are 5 types of modeling functions:
-
_observe: We provide observation data for this component. For example, we can useconcentrations_observe()if we have observed concentration measurements and want to fit the model to them. -
_assume: We assume the values for this component. For example, we can usegeneration_dist_assume()to provide a generation time distribution from the literature. -
_calibrate: This is similar to_assume, but instead of directly specifying the value for an assumption, we calibrate it to some other assumption or data. For example, we can useload_per_case_calibrate()to calibrate the shedding load per case to case data (so that the estimated infections will roughly match the observed case numbers). -
_estimate: We estimate this component as a parameter of the model. For example, we can usenoise_estimate()if we don’t know how much noise the measurements have and want to estimate this from the data. -
_none: We do not model this component. For example, we can usesample_effects_none()if we don’t want to model any sample effects on the measurements.
To find out which modeling functions are available for a given
component, you can consult the documentation or use the helper
component_functions():
EpiSewer::component_functions("infection_noise")
#> [1] "infection_noise_none()" "infection_noise_estimate()"💡 Multiple modeling options
Some components have multiple versions of the same modeling function
type. For example, there are several approaches to estimate the
reproduction number, such as R_estimate_gp() (Gaussian
process), R_estimate_rw() (random walk),
R_estimate_ets() (exponential smoothing),
R_estimate_splines() (smoothing splines) and more.
EpiSewer::component_functions("R")
#> [1] "R_estimate_gp()" "R_estimate_ets()"
#> [3] "R_estimate_smooth_derivative()" "R_estimate_piecewise()"
#> [5] "R_estimate_changepoint_splines()" "R_estimate_approx()"
#> [7] "R_estimate_rw()" "R_estimate_splines()"❗ Modeling restrictions
Not all components support all modeling types. For example,
EpiSewer currently only offers
generation_dist_assume(), but not
generation_dist_estimate,
generation_dist_calibrate, or
generation_dist_observe.
EpiSewer::component_functions("generation_dist")
#> [1] "generation_dist_assume()"This is because estimating the generation time distribution from data is not yet supported (but may be added in the future).
Data and assumptions
The sewer_data() and sewer_assumptions()
functions are convenience functions that allow you to collect all
observation data and modeling assumptions in one place. This can improve
overview and allows to run EpiSewer under its default
settings without repeating the component definitions:
ww_data <- sewer_data(
measurements = SARS_CoV_2_Zurich$measurements,
flows = SARS_CoV_2_Zurich$flows,
cases = SARS_CoV_2_Zurich$cases # optional
)
ww_assumptions <- sewer_assumptions(
generation_dist = get_discrete_gamma_shifted(gamma_mean = 3, gamma_sd = 2.4),
shedding_dist = get_discrete_gamma(gamma_shape = 0.929639, gamma_scale = 7.241397),
shedding_reference = "symptom_onset",
incubation_dist = get_discrete_gamma(gamma_shape = 8.5, gamma_scale = 0.4),
)
EpiSewer(
data = ww_data,
assumptions = ww_assumptions
)What happens under the hood is that when individual model components
are not provided with the data or assumptions they need, they search the
data and assumption arguments passed to the EpiSewer()
function.
💡 It is always possible to mix both approaches and specify data or assumptions explicitly in the model component:
# Leave out flows from the data
ww_data <- sewer_data(
measurements = SARS_CoV_2_Zurich$measurements,
#flows = SARS_CoV_2_Zurich$flows
cases = SARS_CoV_2_Zurich$cases
)
# Leave out the generation time distribution
ww_assumptions <- sewer_assumptions(
#generation_dist = get_discrete_gamma_shifted(gamma_mean = 3, gamma_sd = 2.4),
shedding_dist = get_discrete_gamma(gamma_shape = 0.929639, gamma_scale = 7.241397),
shedding_reference = "symptom_onset",
incubation_dist = get_discrete_gamma(gamma_shape = 8.5, gamma_scale = 0.4),
)
# Provide flows directly to sewage module
ww_sewage <- model_sewage(
flows = flows_observe(SARS_CoV_2_Zurich$flows)
)
# Provide generation time distribution directly to infections module
ww_infections <- model_infections(
generation_dist = generation_dist_assume(
get_discrete_gamma_shifted(gamma_mean = 3, gamma_sd = 2.4)
)
)
# Combine everything
result <- EpiSewer(
data = ww_data,
assumptions = ww_assumptions,
sewage = ww_sewage,
infections = ww_infections
)Note that if the same data or assumptions are supplied via the
sewer_data()/sewer_assumptions() and
the individual component, EpiSewer() will compare both
arguments and throw an error if they differ.